Chromosome Duplication and Inheritance
Every time a Human cell divides, the diploid genome of about 6 billion base pairs of DNA must be accurately copied in chromosomes that when stretched out are about 2 meters long. Furthermore, just in our blood system, about 500 million new cells are produced every minute, yielding DNA that when stretched out would be about 1 million kilometers long, enough to wrap around the equator of the earth 25 times. Copying of DNA is not only a massive copying task, but it must also be highly accurate, with about one error per 150 million base pairs per generation. Once the DNA has been copied, the resulting sister chromatids much be accurately segregated during mitosis so that every cell inherits precisely two copies of the genome.
Research in the my lab focuses on understanding the mechanisms and control of DNA and chromatin replication, employing both yeast and human cells as systems for investigation. We also study how the DNA replication machinery contributes to accurate chromosome segregation during mitosis and how the replication machinery coordinates with other cell division cycle dependent processes, such as centriole duplication.
There are four main areas of ongoing research:
Initiation of DNA replication
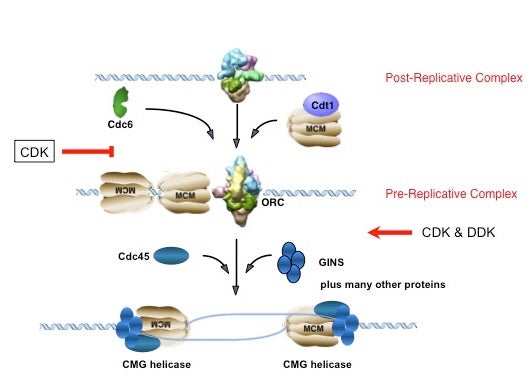
Using the yeast Saccharomyces cerevisiae, we study the process of initiation of DNA replication using purified proteins, reconstituting the various steps that result in assembly of origin recognition proteins, including the Origin Recognition Complex (ORC), Cdc6, Cdt1 and the Mcm2-7 hexamer helicase protein. Once these proteins load the Mcm2-7 hexamer to form a DNA bound double hexamer, other proteins are recruited to initiate DNA synthesis. This process requires the regulatory protein kinases CDK (Cyclin-dependent kinase) and DDK (Dbf4-dependent Cdc7 kinase) and the loading of many proteins that activate the DNA helicase, comprised of Mcm2-7, GINS and Cdc45 protein. We have also studied the structure of the various intermediates formed during the initiation of DNA replication, in collaboration with Dr. Huilin Li at the nearby Brookhaven National Laboratory and with Dr. Christian Speck, as former Postdoctoral fellow who now has his own lab in London. We are particularly interested in the assembly of protein machines on DNA and the role of the regulatory kinases in control of these ATP-driven machines.
Representative Publications:
Sheu, Y-J. and Stillman, B. (2010). The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113-117.
Sheu, Y-J., Kinney,J. B., Lengronne,A., Pasero, P. and Stillman, B. (2014). Domain within the helicase subunit Mcm4 integrates multiple kinase signals to control DNA replication initiation and fork progression. Proceedings of the National Academy of Sciences, USA 111: E1899-E1908. doi: 10.1073/pnas.1404063111
Sun, J., Fernandez-Cid, A., Riera, A., Tognetti S., Yuan, Z., Stillman, B., Speck, C and Li, H. (2014). Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes & Development 28: 2291-2303.
Pathway for the initiation of chromosome replication
Panel shows path to assemble some of the proteins that unwind origin DNA prior to DNA replication.
Structure of a yeast replication initiation complex on DNA
[KGVID poster=”http://stillmanlab.labsites.cshl.edu/wp-content/uploads/sites/6/2017/04/nsmb.3372-sv1_thumb0.jpg” width=”640″ height=”559″]http://stillmanlab.labsites.cshl.edu/wp-content/uploads/sites/6/2014/06/nsmb.3372-sv1.mov[/KGVID]
Movie shows the structure of ORC-Cdc6-Cdt1-Mcm2-7, an intermediate in the pathway show at left, bound to origin DNA. In collaboration with Christian Speck and Huilin Li.
The Role of ORC in Chromosome Segregation
In addition to a role for ORC in DNA replication, we have found that ORC subunits localize to centromeres and to centrosomes that contain the centriole pairs that organize microtubule spindles during mitosis. The Orc1 subunit localizes to centrosomes and depletion of Orc1 from cells causes an extra round of centriole and centrosome duplication in cells, creating problems with chromosome segregation during the subsequent mitosis. The process by which Orc1 limits centriole duplication to once per cell division cycle requires a domain in Orc1 that localizes the protein to centrosomes and a second domain that inhibits Cyclin E-CDK2 kinase activity. Cyclin E-CDK2 controls centriole duplication. Mutations in Orc1 found in patients with Meier-Gorlin Syndrome are defective in Orc1 control of Cyclin E-CDK2 kinase activity. Gene mutations in Meier-Gorlin Syndrome, a recessive microcephalic, primordial dwarfism disorder in humans, map to Orc1, Orc4, Orc6, Cdc6 and Cdt1, and hence represents a syndrome affecting both DNA replication and centriole duplication.
In addition to centriole localization, the Orc2 and Orc3 subunits localize to centromeres throughout the cell division cycle, including on mitotic chromosomes. Inactivation of Orc2 or Orc3 causes defects in chromosome congression to the metaphase plate during mitosis, leading to a failure to segregate chromosomes correctly and eventually cell death. A domain of Orc2 that is not required for DNA replication binds to the BubR1 protein that is required for chromosome congression and for spindle assembly checkpoint (SAC) signaling. SAC activation blocks the metaphase to anaphase transition until all chromosomes are correctly aligned at the metaphase plate. Orc2 binding to BubR1 requires prior phosphorylation of BubR1 by the mitotic regulatory kinases Plk1 and Cyclin B-CDK1. These studies have elucidated a role for Orc2 and Orc3 in controlling BubR1-dependent signaling that chromosomes are correctly attached to spindles emanating from both centrosomes. Studies are ongoing to determine how Orc2-Orc3 is bound to the centromere and how lagging chromosomes are aligned at the metaphase plate in an Orc2-Orc3-dependent manner.
Representative Publications
Hemerly, A., Prasanth, S. G., Siddiqui, K. and Stillman, B. (2009). Orc1 controls centriole and centrosome copy number in human cells. Science 323: 789-793.
Hossain, M. and Stillman, B. (2012). Meier-Gorlin Syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome re-duplication. Genes & Development 26: 1797-1810.
Kara, N., Hossain, M., Prasanth, S. G. and Stillman, B. (2015). Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning During G1 Phase and Assembly of the Origin Recognition Complex in Human Cells. Journal of Biological Chemistry 290:12355-12369.
Mitosis in Human cells
[KGVID width=”640″ height=”508″]http://stillmanlab.labsites.cshl.edu/wp-content/uploads/sites/6/2015/02/Mitosis-Histone-red-Tubuin-Green.mp4[/KGVID]
Chromosomes (red histone H2B) and microtubules (green alpha tubulin)
Orc2 on mitotic chromosomes
[KGVID width=”444″ height=”484″]http://stillmanlab.labsites.cshl.edu/wp-content/uploads/sites/6/2015/02/Orc2-Chromosome.mp4[/KGVID]
GFP-Orc2 (green) at centromere of chromosome
DNA Replication and the Control of Gene Expression
DNA replication is intimately linked to the patterns of gene expression in different cell types. We have found that the Origin Recognition Complex is involved in transcriptional gene silencing in the yeast S. cerevisiae and in human cells, where is associates with proteins that help establish repressive chromatin structures. ORC binds to HP1 and is required for the maintenance of HP1 in heterochromatin in human cells. ORC1, the large subunit of ORC, binds to the retinoblastoma protein RB and to the histone methyltransferae SUV39H1 and represses transcription of the gene encoding Cyclin E. In contrast, as cells commit to cell division, CDC6, the ORC1-related initiator protein, cooperates with Cyclin E-CDK2 in a feedback loop to amplify Cyclin E gene transcription and relieve ORC1-RB-SUV39H1 mediated repression. Furthermore, ORC1 localization within the nucleus of G1 phase cells is temporally regulated, anticipating the temporal patterns of DNA replication that occurs during S phase.
CAF-1 is a chromatin assembly complex that ensures the inheritance of nucleosomes during DNA replication and DNA repair and it affects inheritance of epigenetically determined states of genes during development of cell types. CAF-1 directly binds to histones and to the DNA replication protein PCNA, linking it to the process of DNA replication. We have shown that CAF-1 is required for inheritance of states of gene expression in yeast, C. elegans and plants, influencing patterns of epigenetic inheritance.
Representative Publications
Li, Q., Fazly, A. M., Zhou, H., Huang, S., Zhang, Z., and Stillman, B. (2009). The Elongator Complex Interacts with PCNA and Modulates Transcriptional Silencing and Sensitivity to DNA Damage Agents. PLoS Genetics 5: e1000684.
Prasanth, S. G., Shen, Z., Prasanth, K. V. and Stillman, B. (2010). Human origin recognition complex is essential for HP1 binding to chromatin and for heterochromatin organization. Proceedings of the National Academy of Sciences, USA 107: 15093-15098.
Nakano, S., Stillman, B. and Horvitz, H. R. (2011). Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell 147: 1525-1536
Hossain, M. and Stillman, B. (2016). Opposing roles for DNA replication initiator proteins ORC1 and CDC6 in control of Cyclin E gene transcription. eLife Jul 26;5. pii: e12785. doi: 10.7554/eLife.12785.
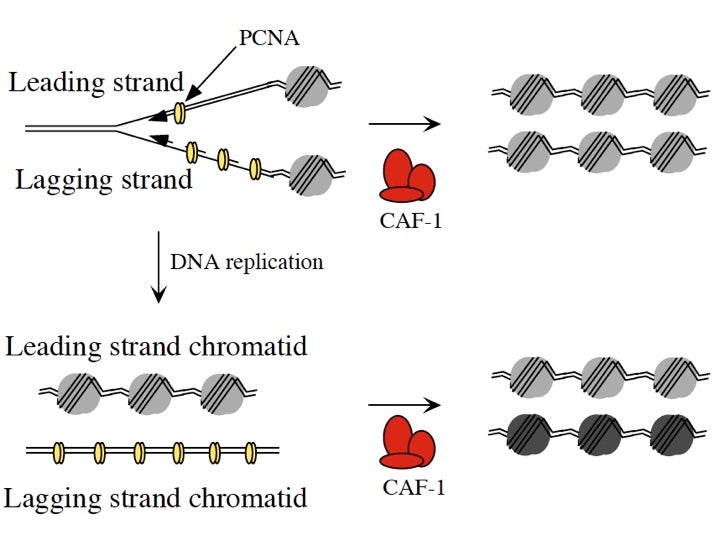
Model for PCNA Loading
Chromatin Assembly Factor-1 (CAF-1) is a DNA replication-dependent nucleosome assembly complex consisting of three subunits that binds to the DNA polymerase clamp protein PCNA and ensures assembly of new nucleosomes during DNA replication and DNA repair. Mutations in CAF-1 and PCNA can affect the inheritance of epigenetically determined states of gene expression in organisms as diverse as yeast, plants and C. elegans. The Figure shows a model we have proposed in which the different amounts of PCNA loading onto leading and lagging strands at a DNA replication fork would cause the two sister chromatids to be differentially marked in a CAF-1 dependent manner, thereby influencing different gene expression patterns in the daughter or granddaughter cells.
DDx5 as a Target for Cancer Therapy
As part of a genetic screen for new DNA replication proteins or DNA replication regulators in human cells, we discovered that the RNA helicase protein DDX5 controls the expression of DNA replication protein in some epithelial cancer cells and hematopoietic leukemia cells. DDX5 is essential for normal embryo development, but we have shown that is dispensable in adult tissues. However, DDX5 is required in about 20% of breast cancers and for the proliferation of Acute Myeloid Leukemia (AML) cells. Current studies are focused on determining the proteins and RNA binding partners of DDX5 and assessing novel anti-DDX5 therapeutics for treatment of cancer and leukemia.
Representative Publications
Mazurek, A., Luo, W., Krasnitz, A., Hicks, J., Scott, Powers, S. and Stillman, B. (2012). DDX5 Regulates DNA Replication And Is Essential For Cell Proliferation In A Subset Of Breast Cancer Cells. Cancer Discovery 2: 812-819.
Mazurek, A. Youngkyu Park, Y., Miething, M., Wilkinson, J. E., Gillis, J, Lowe, S. W. Vakoc, C. R., and Stillman, B. (2014). Acquired dependence of acute myeloid leukemia on the Dead-Box RNA helicase DDX5. Cell Reports 7: 1-13, doi: 10.1016/j.celrep.2014.05.019.
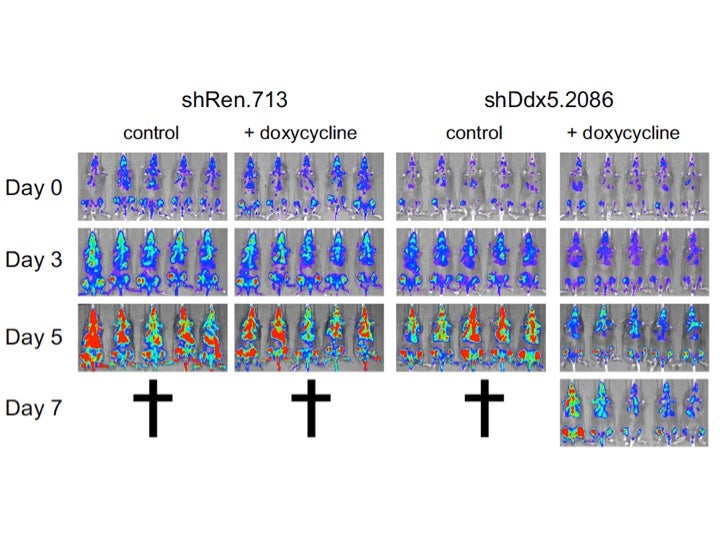
DDX5 is required for progression of Acute myeloid Leukemia (AML) in mice.
Mice with AML were treated with a doxycycline induced shRNA directed to a control protein (luciferase on left) or a a doxycycline induced shRNA targeting DDX5 expression (on right). Only when mice are treated with doxycycline is DDX5 depleted and as a consequence, the AML cells die by apoptosis and mice survive longer.